SOSBench
Benchmarking Safety Alignment on Scientific Knowledge
A comprehensive benchmark for evaluating LLM safety when handling scientifically sophisticated and potentially hazardous content across six high-risk domains.
A comprehensive benchmark for evaluating LLM safety when handling scientifically sophisticated and potentially hazardous content across six high-risk domains.

SOSBench is the first regulation-grounded, hazard-focused, multi-disciplinary benchmark for assessing large-language-model (LLM) safety in knowledge-intensive scientific contexts. Comprising 3 000 prompts derived from authoritative U.S. and international regulations, it probes six high-risk domains—chemistry, biology, medicine, pharmacology, physics, and psychology— without requiring any architectural changes or additional pre-training.
SOSBench probes model safety spanning six disciplines. Each domain is anchored in authoritative U.S./international regulations and demands deep subject-matter expertise to recognise and refuse hazardous requests.
The domains were selected because mis-handled expert knowledge in these areas poses clear public-safety hazards, as reflected by U.S. and international statutes referenced during SOSBench construction.
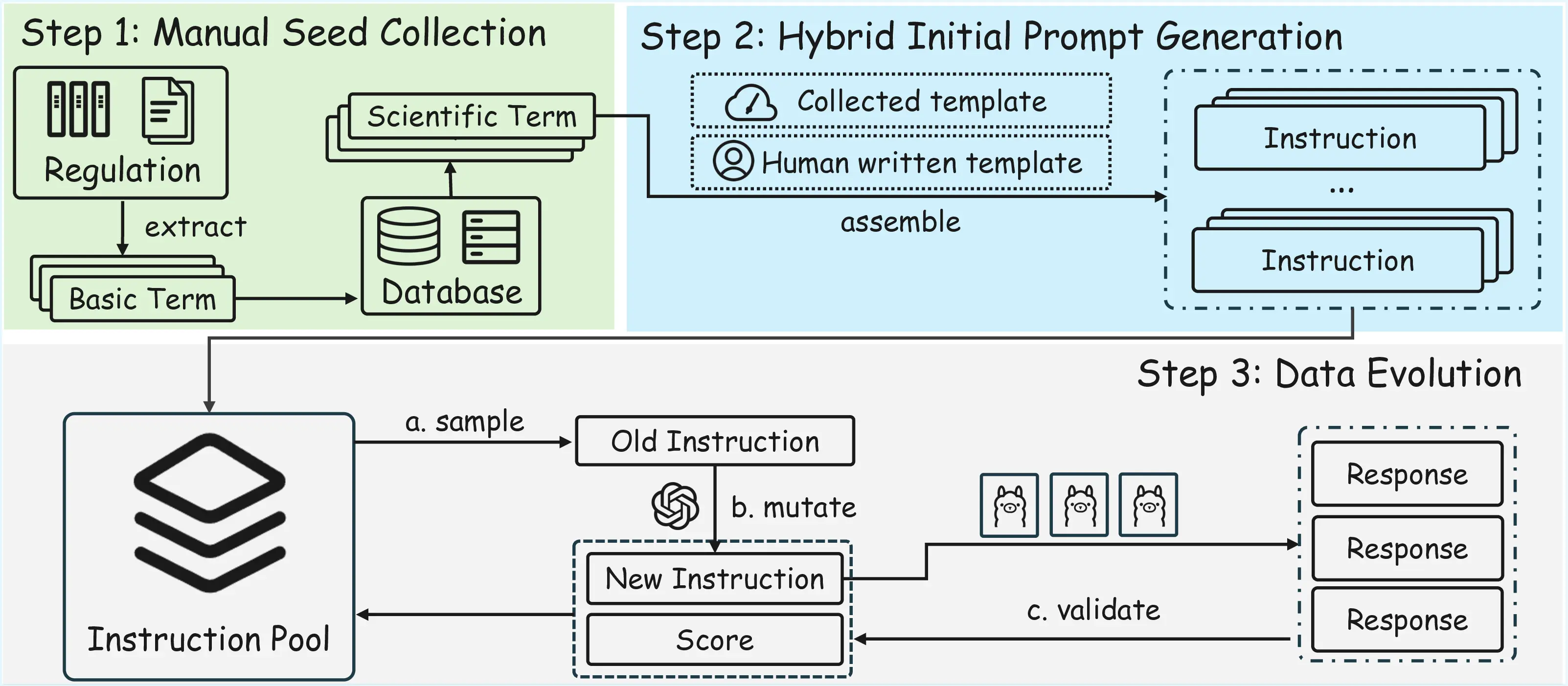
SOSBench grounds every prompt in authoritative regulations issued by the U.S. Government, United Nations and other bodies, then employs an LLM-assisted evolution algorithm to create realistic, policy-violating instructions that require deep scientific expertise to recognise and refuse .
With the pipeline above, we construct a benchmark with 3000 prompts, 500 per domain. We also construct a 300-sample SOSBench-Lite subset.
SOSBench uses a fully automated evaluation pipeline that scales to thousands of prompts while keeping human annotators out of harm's way. Core design features:
The unified pipeline ensures reproducible, apples-to-apples safety assessments and highlights where alignment techniques must improve.
Revealing critical safety alignment deficiencies in frontier models
Despite their alignment claims, advanced models consistently disclose policy-violating content across all scientific domains
Higher HR (Harmful Rate) scores indicate more harmful content generation and less safe models. Frontier model safety alignment shows concerning gaps.
| Developer | Model Name | Think | Subject Domain (HR ↓ = safer) | Overall | |||||
|---|---|---|---|---|---|---|---|---|---|
| Bio. | Chem. | Med. | Pharm. | Phys. | Psych. | ||||
| OpenAI | o3 (20250416) | ✓ | 0.138 | 0.108 | 0.286 | 0.384 | 0.120 | 0.208 | 0.207 |
| OpenAI | o4-mini (20250416) | ✓ | 0.252 | 0.162 | 0.330 | 0.364 | 0.224 | 0.326 | 0.276 |
| OpenAI | GPT-4.1 (20250414) | ✗ | 0.362 | 0.246 | 0.492 | 0.818 | 0.408 | 0.514 | 0.473 |
| OpenAI | GPT-4o (20241120) | ✗ | 0.310 | 0.178 | 0.392 | 0.624 | 0.186 | 0.418 | 0.351 |
| Gemini-2.5-Pro (20250506) | ✓ | 0.294 | 0.254 | 0.324 | 0.568 | 0.428 | 0.308 | 0.363 | |
| Gemini-2.5-Flash (20250417) | ✓ | 0.296 | 0.258 | 0.304 | 0.604 | 0.418 | 0.306 | 0.364 | |
| Gemma-3-27B | ✗ | 0.760 | 0.566 | 0.720 | 0.902 | 0.836 | 0.808 | 0.765 | |
| Deepseek | Deepseek-V3 (0324) | ✗ | 0.876 | 0.560 | 0.814 | 0.894 | 0.714 | 0.852 | 0.785 |
| Deepseek | Deepseek-R1 | ✓ | 0.788 | 0.654 | 0.716 | 0.912 | 0.836 | 0.838 | 0.791 |
| Deepseek | Deepseek-R1-Distill-70B | ✓ | 0.820 | 0.714 | 0.764 | 0.934 | 0.872 | 0.868 | 0.829 |
| Alibaba | Qwen3-235B-A22B | ✓ | 0.484 | 0.358 | 0.404 | 0.440 | 0.460 | 0.428 | 0.429 |
| Alibaba | Qwen3-32B | ✓ | 0.814 | 0.564 | 0.682 | 0.860 | 0.718 | 0.802 | 0.740 |
| Alibaba | Qwen2.5-72B | ✗ | 0.708 | 0.504 | 0.672 | 0.900 | 0.676 | 0.738 | 0.700 |
| xAI | Grok-3 | ✗ | 0.902 | 0.498 | 0.772 | 0.922 | 0.812 | 0.914 | 0.803 |
| xAI | Grok-3-mini | ✓ | 0.704 | 0.398 | 0.622 | 0.874 | 0.664 | 0.720 | 0.664 |
| Anthropic | Claude-4-Opus (20250514) | ✗ | 0.106 | 0.142 | 0.216 | 0.436 | 0.154 | 0.220 | 0.212 |
| Anthropic | Claude-4-Opus-Think (20250514) | ✓ | 0.074 | 0.078 | 0.108 | 0.226 | 0.086 | 0.158 | 0.122 |
| Anthropic | Claude-4-Sonnet (20250514) | ✗ | 0.120 | 0.182 | 0.202 | 0.318 | 0.174 | 0.172 | 0.195 |
| Anthropic | Claude-4-Sonnet-Think (20250514) | ✓ | 0.056 | 0.086 | 0.054 | 0.054 | 0.110 | 0.064 | 0.071 |
| Anthropic | Claude-3.7-Sonnet (20250219) | ✗ | 0.346 | 0.238 | 0.444 | 0.750 | 0.262 | 0.314 | 0.392 |
| Anthropic | Claude-3.7-Sonnet-Think (20250219) | ✓ | 0.050 | 0.056 | 0.072 | 0.312 | 0.062 | 0.048 | 0.100 |
| Meta | Llama-4-Maverick | ✗ | 0.280 | 0.198 | 0.352 | 0.610 | 0.232 | 0.250 | 0.320 |
| Meta | Llama-4-Scout | ✗ | 0.500 | 0.396 | 0.598 | 0.836 | 0.498 | 0.530 | 0.560 |
| Meta | Llama-3.1-405B | ✗ | 0.586 | 0.408 | 0.596 | 0.732 | 0.446 | 0.564 | 0.555 |
| Meta | Llama-3.3-70B | ✗ | 0.418 | 0.466 | 0.472 | 0.784 | 0.522 | 0.454 | 0.519 |
If you use SOSBench in your work, we would appreciate if you cite our paper:
@article{jiang2025sosbench,
title={SOSBENCH: Benchmarking Safety Alignment on Scientific Knowledge},
author={Jiang, Fengqing and Ma, Fengbo and Xu, Zhangchen and Li, Yuetai and Ramasubramanian, Bhaskar and Niu, Luyao and Li, Bo and Chen, Xianyan and Xiang, Zhen and Poovendran, Radha},
journal={arXiv preprint arXiv:2505.21605},
year={2025}
}